Research
Why lncRNA?
The genomes of all organisms store the information necessary for proper development and survival. Contrary to initial assumptions, eukaryotic genomes are not dominated by protein-coding genes but are composed of 98% of non-coding regions, also called “dark matter”. Undoubtedly, it can be said that this dark DNA and the non-coding RNAs produced from it are the rising stars of the twenty-first century and a major focus of genomic studies. Long noncoding RNAs (lncRNAs) – long (>200 nucleotides) RNA molecules with limited protein-coding potential constitute the biggest yet most enigmatic class of noncoding RNAs. Once considered transcriptional noise, lncRNAs are now being recognized as molecules with critical regulatory functions.
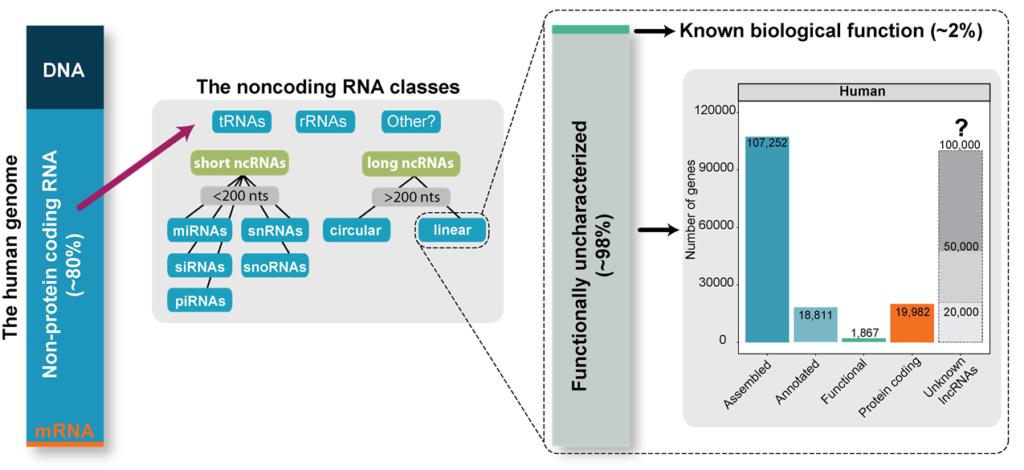
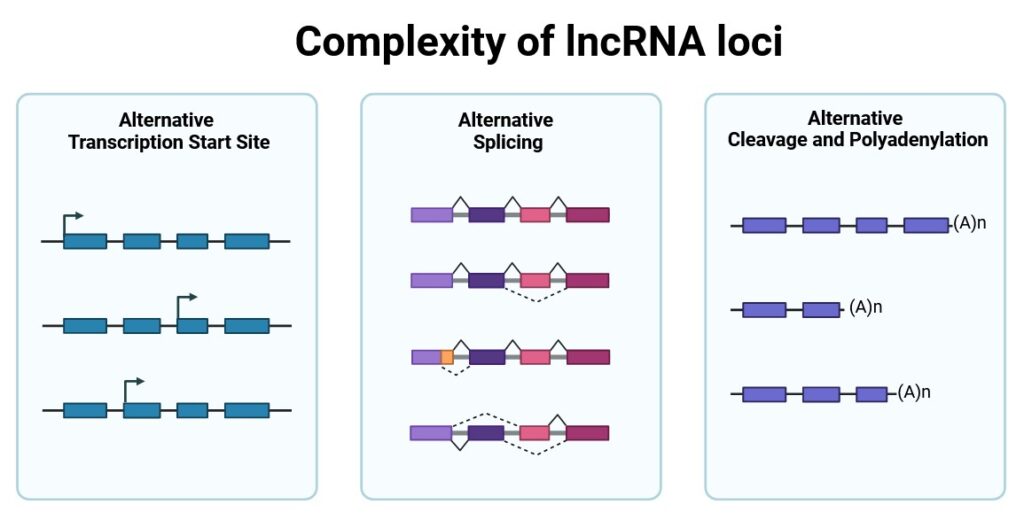
A constantly growing number of lncRNAs are being associated with diverse roles in crucial biological processes or disease progression. Moreover, the highly specific spatio-temporal expression patterns of lncRNAs make them perfect biomarkers or therapeutic targets, which further confirms their high biomedical significance and creates the need for their in-depth understanding. Unfortunately, so far, only <3% of ~100,000 lncRNA genes encoded by the human genome have been functionally characterized, leaving thousands of biomedically relevant genes undiscovered. Moreover, despite extensive studies, we still lack the basis to confidently state which lncRNAs are functional and how lncRNA functions are preserved in their primary RNA sequence. Therefore, all efforts are directed towards in-depth exploration of lncRNAs, including the identification of sequence-structure-function relationships and studying their evolutionary conservation and regulatory aspects of their expression. It is believed that a better understanding of lncRNA biology will facilitate their proper characterization and yield with discovery of numerous functional genes.
LncRNAs in zebrafish
Understanding lncRNA relevance in the context of human biology and disease can be facilitated by the application of animal models. Zebrafish (Danio rerio) is a fully developed vertebrate organism that appears to be a powerful system for studying essential developmental and cellular processes, as well as human pathogenesis. Physiological and anatomical similarities (analogous major organs and tissues) together with the conservation of 70% of genes between human and zebrafish ensure that information acquired through danio rerio is more accurate than that obtained by in vitro studies or in non-vertebrate organisms. Hence, zebrafish is an ideal model to provide insights into lncRNA genomics, evolution, regulation and function.
However, recent zebrafish lncRNA annotations lag considerably behind those for human and mouse in terms of both detected gene loci and the quality of existing lncRNA models. Another aspect that is hampering zebrafish usage is the difference in annotation bias, which makes it difficult to directly compare human and zebrafish lncRNA annotations. Even more importantly, contrary to mRNAs lncRNAs display lower evolutionary conservation and low expression levels, which significantly impede their identification and biological characterization.
The goal of our lab is to establish innovative bioinformatic strategies to explore evolutionary conservation of lncRNAs in distant species, to optimize the experimental protocols for lncRNA catalog improvement, and all together to facilitate lncRNA functional characterization studies.
Next, we aim to use these strategies to produce a high-quality lncRNA annotation for the zebrafish genome by improving identification of lncRNA orthologs and removing annotation bias toward developmental samples and, as a result, to expand zebrafish application as an animal model to study lncRNA functions.
Post-transcriptional processing of non-coding RNA during early embryogenesis
Every complex multicellular organism is formed from a single-celled zygote through a series of strictly controlled developmental processes. Each of these steps requires the precise temporal and spatial expression of specific genes, which is ensured by the fine-tuned balance between RNA synthesis and degradation. Interestingly, specific post-transcriptional modifications such as capping, splicing and polyadenylation affect the stability of mature RNA molecules and help determine their cellular fate. So far, the greatest emphasis in transcriptomic studies has been placed on mRNAs while mostly neglecting other RNA molecules from the analysis.Therefore, we still do not fully understand the principles of post-transcriptional modifications of non-coding RNA. This is particularly true for lncRNAs which tend to have lower expression, splicing efficiency and stability with more restricted developmental and tissue-specific expression patterns compared to mRNAs.
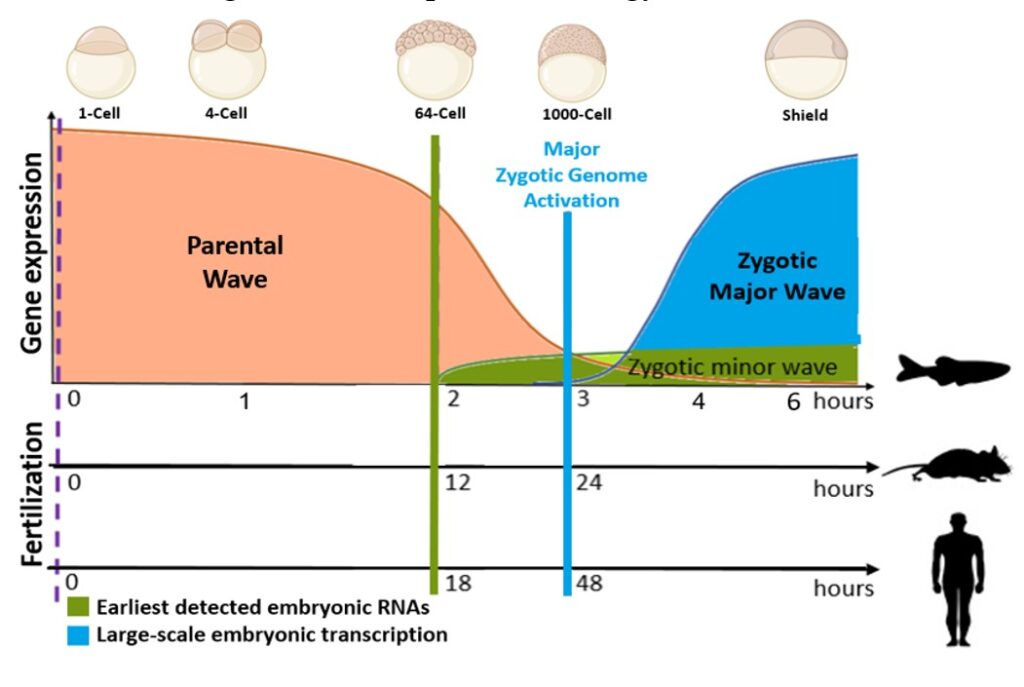
Our goal is to explore the regulation mechanisms of the non-coding part of the transcriptome during the Maternal-to-Zygotic Transition (MZT) in zebrafish, with particular focus on lncRNAs. The MZT is a complex process occurring during early embryogenesis in higher plants and animals that defies the exact timing of Zygotic Genome Activation (ZGA). As the MZT encompass two processes: (1) the degradation of parentally inherited transcripts (and proteins) deposited in the egg cell and (2) the synthesis of the very first zygotic transcripts (ZGA), it is an ideal model for studying mechanisms of post-transcriptional processing of lncRNAs.